Overview
Gene imprinting is the expression of certain genes depending on their parent-of-origin, essential for normal development in both flowering plants and placental mammals. In plants, imprinting is mostly confined to the endosperm of the developing seed. DNA demethylation of specific loci in the female gametophyte is central to the control of this epigenetically regulated process. As such, DNA demethylation is essential during plant development due to its role in the regulation of gene imprinting. Our recent data, together with our collaborators in Daniel Zilberman’s lab, support this, but also reveal a more basal function for this process. DNA demethylation by DME seems to primarily regulate the activity of transposable elements, in order to protect the genomic integrity of progeny. The monoallelic silencing of genes whose expression is controlled by TEs, if conferring a selection advantage, may have led to the evolution of imprinted loci in flowering plant endosperm.
Double fertilisation
The female Arabidopsis gametophyte is formed within the ovule. A haploid spore undergoes three mitotic divisions to form an 8-nucleus, 7-cell female gametophyte containing the egg, central, synergid, and antipodal cells. Before fertilization, a diploid nucleus is formed in the central cell by the fusion of two haploid nuclei. The endosperm is derived from fertilization of the central cell by a sperm cell, whilst fertilization of the adjacent egg cell by a second sperm cell gives rise to the embryo. Thus, double fertilization generates a seed with a triploid endosperm and diploid embryo.
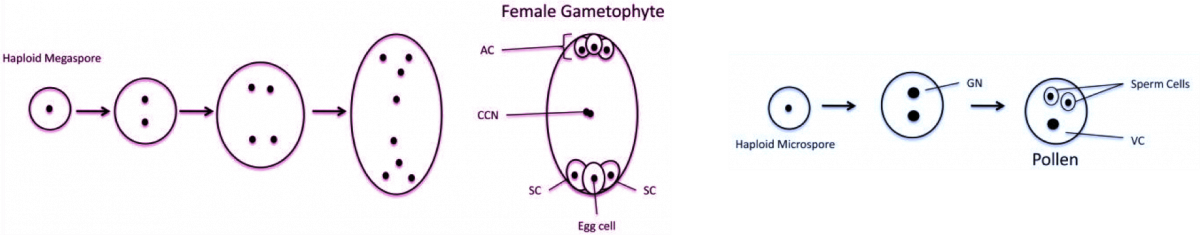
The female and male gametophytes in Arabidopsis, From Bauer and Fischer., 2011; PDF; AC, Antipodal cells; CCN, Central Cell Nucleus; SC, Synergid Cell; GN, Generative nucleus; VC, Vegetative Cell.
Using a screen to identify regulators of seed development, we found that the DEMETER (DME) gene is required for seed development. DME encodes a large protein with multiple conserved domains, including DNA glycosylase and nuclear localization domains (Choi et al., 2002; PDF, Mok et al., 2010; PDF). A maternal DME allele is required for seed viability, and maternal inheritance of a mutation results in 50 % seed abortion (see below) (Choi et al., 2002; PDF). DME is a demethylase, able to excise 5-methylcytosine in vitro via the Base Excision DNA repair pathway (Gehring et al., 2006; PDF).
Gene Imprinting
Gene imprinting is a fascinating process, absolutely required for normal development in flowering plants and placental mammals. It seems to function in resource allocation, batting firstly for the paternal side, allowing the development of an organ whose purpose is to channel nutrients to developing offspring (the placenta, or endosperm in seeds), ensuring the maximum inheritance of the paternal genome. In order for the mother to also maximize propagation of her own genome, she must spread those resources across all of her offspring, including those from future reproductive cycles. Maternally expressed imprinted genes therefore limit the resources she provides, whereas paternally expressed genes do the opposite. The discovery of imprinting in mammals and the theory of conflict are descibed by the following papers:Barton et al., 1984; PDF, McGrath and Solter, 1984; PDF, Moore and Haig, 1991; PDF.
In both plants and mammals DNA methylation (5-methylcytosine) is a critical regulator of gene imprinting. In Arabidopsis, DNA demethylation is a key regulator of imprinting and the action of DME is required for parent-specific expression of many genes (Hsieh et al., 2011; PDF). One important example is the regulation of MEDEA, another critical gene for seed development whose locus is capable of autonomous regulation. The MEDEA (MEA) gene is named after a goddess in Greek mythology who killed her children (by accident, or intentionally, depending on which myth you read). The MEA protein is an essential subunit of the reproductive Polycomb group repressive complex (PRC2), and its expression is imprinted in the endosperm. DME is responsible for endosperm maternal-allele-specific hypomethylation at the MEA gene Gehring et al., 2006; PDF) DME specifically excises 5-methylcytosine at the MEA promoter, activating its transcription in the central cell of the female gametophyte. After fertilization of the central cell, the maternal expression of MEA results in PRC2-mediated silencing of its own paternal allele, and additional imprinted genes in the endosperm.
The endosperm in Arabidopsis is a unique tissue; not only does it harbour the majority of imprinted transcripts, but it has a different DNA methylation profile compared to other tissues. Following genome-wide analysis of DNA methylation in the endosperm and embryo of developing seeds, we found the endosperm to exhibit global hypomethylation when compared to the embryo. This hypomethylation is independent of DME, and so far its function is unknown (Hsieh et al., 2009; PDF). Subsequent to these analyses, we investigated gene imprinting in Arabidosis endosperm genome-wide. Prior to the advent of Next Generation Sequencing, only nine genes in A. thaliana were known to be imprinted. In our 2011 paper in PNAS, Hsieh et al., we found an additional 120 maternally expressed imprinted genes and 9 paternally expressed imprinted genes. Similarly to mammals, the regulation of imprinting is complex, we find canonical regulatory mechanisms at play – the DNA demethylation of maternally derived promoter regions leading to maternal specific expression; paternal methyltransferase-mediated methylation of polycomb-group binding targets leading to paternal specific expression; as well as more complex regularity mechanisms, yet to be delineated. The genes that we found to be imprinted contribute to a wide variety of processes, many contributing to epigenetic regulation themselves.
A common role for DME in the male and female gametophytes
The maternal alleles of the endosperm; likely reflecting the central cell genome; and the genome of the vegetative cell, are hypomethylated compared to that of the embryo or sperm. This hypomethylation is dependent on DME, occurs in the CG, CHG and CHH DNA contexts and is enriched at short, AT rich transposon sequences. DME is also expressed in the male gametophyte, in the vegetative cell, as established by careful experiments to isolate sperm and vegetative cells using FACS, in the lab of our collaborator Hisashi Tamaru. In the vegetative cell, DME carries out a similar program of DNA demethylation to that in the female gametophyte. Since the vegetative cell does not contribute genetic material to the developing seed, the effect of a loss of DME is more subtle, manifesting as a reduced transmission of paternal dme, in certain ecotypes, due, at least in part, to reduced pollen germination (Schoft et al., 2011; PDF).
These regions of DNA demethylation are often associated with the differentially methylated regions controlling gene imprinting. 45 % of hypomethylated loci in the vegetative cell and endosperm are overlapping, highly suggestive of a parallel process occurring in the vegetative and central “companion” cells. It is thought that TE expression in companion cells generates small RNAs that reinforce silencing of those same TE sequences in gametes, protecting their genomes them from potentially deleterious transposition (Ibarra et al., 2012; PDF).
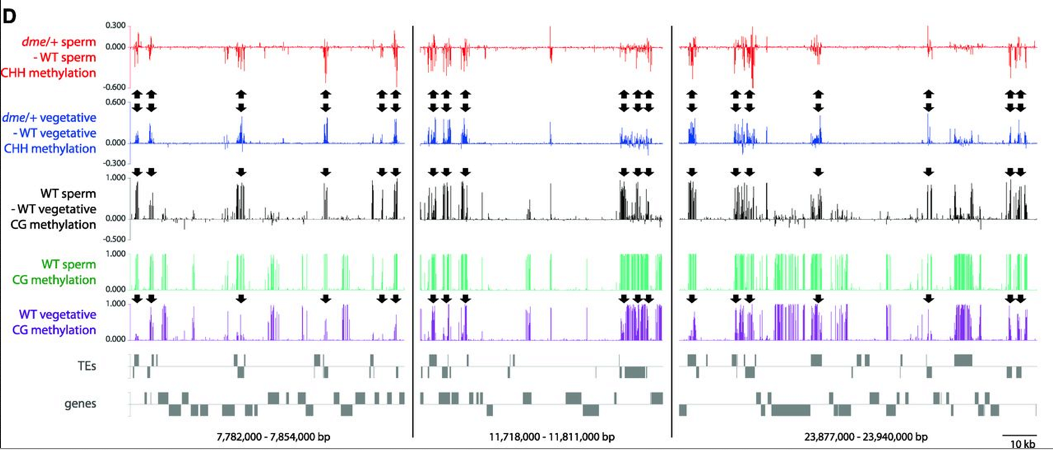
CG and CHH methylation in pollen on chromosome 1. Arrows point out loci with decreased CHH methylation in sperm and increased CG methylation in vegetative cells from dme/+ pollen (Ibarra et al., 2012)
In support of this hypothesis, an artificial miRNA directed at GFP and expressed specifically in the central cell, was able to silence GFP expression in the egg cell. Further, demonstrating that silencing induced by companion cell sRNAs likely occurs at the transcriptional level, i.e. by RNA directed DNA methylation (RdDM), lack of DME in the companion cell seems to reduce RdDM at DME target sequences in gametes. RdDM confers CHH methylation at sites dictated by the sequence specificity of 24nt siRNAs. The CG sites that are demethylated by DME in vegetative cells show increased CG methylation in dme mutant vegetative cells. These same loci show preferential CHH hypomethylation in dme mutant sperm (Ibarra et al., 2012).

An anti-GFP microRNA expressed in the central cell correlates with a reduction of GFP expression in the egg (image by Rie Uzawa, from Ibarra et al., 2012).
Demethylation profiles in rice
Rice is a monocotyledonous plant, diverging from the last common ancestor of monocots and dicots 150 million years ago. The rice endosperm persists, unlike that of A. thaliana, and is the dominant staple food crop for approximately half the world’s population. Rice endosperm is hypomethylated in CG, CHG and CHH sequence contexts, compared to the embryo. Our 2013 PNAS paper, Rodrigues et al., (PDF) demonstrates that hypomethylation of the rice endosperm is restricted to the maternal allele, often associated with the control regions of imprinted genes, and tends to occur in euchromatin. A class of imprinted 24 nt sRNAs were found in rice, generated by both parental chromosomes. Intriguingly the loci from which they are expressed correlates with those demethylated on the maternal genome. This sRNA population in rice endosperm spans a wide range of sequences, including TE sequences that tend to be underrepresented in other tissues, supporting the hypothesis that a small RNA pathway exists that upregulates TE expression in companion cells; represented here by the maternal alleles of endosperm tissue; in order to silence TEs in the gametes, as predicted in A.thaliana.

